Current approaches to profiling tissue-specific gene expression in C. elegans require delicate manipulation and are difficult under certain conditions, e.g. dauer or aged worms. Researchers from Tsinghua University have developed a simple and robust method for tissue-specific RNA sequencing by taking advantage of the endogenous trans-splicing process. In this method, transgenic worms are generated in which a spliced leader (SL) RNA gene is fused to a sequence tag and is driven by a tissue-specific promoter. Only in the tissue of interest, the labeled SL RNA gene is transcribed and then trans-spliced into mRNA. The tag allows for the enrichment and sequencing of mRNA from that tissue only.
As a proof of principle, the researchers profiled the muscle transcriptome, which showed high coverage and efficient enrichment of muscle-specific genes, with low background noise. To demonstrate the robustness of the method, they profiled muscle gene expression in aged dauer larvae and worms, revealing changes in gene expression consistent with the physiology of these stages. The resulting muscle transcriptome also revealed 461 new RNA transcripts, probably long noncoding RNAs expressed in muscles.
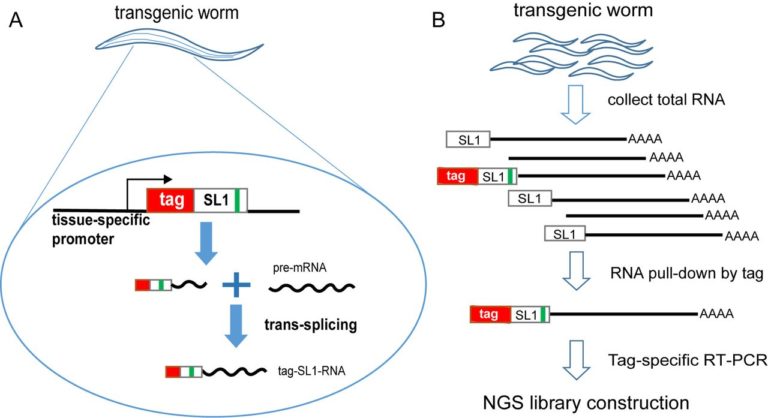
(A) The worm strain carries a sequence-tagged SL1 transgene driven by a tissue-specific promoter. Only in this tissue, the transgenic tag::SL1 will express and trans-splice onto pre-mRNA, resulting in the tag::SL1 fused to the 5′ end of mRNAs. The green bar represents the G16C point mutation introduced to SL1 to distinguish from endogenous wild-type SL1. (B) Flowchart of sequencing library construction. After RNAs were purified from a large number of transgenic worms, those from the tissue of interest were pulled down using the tag sequence as bait. These enriched RNAs are reverse transcribed using polyT primer and polymerase chain reaction (PCR) amplified using the tag as a primer. The tag-based PCR further enriches mRNAs from the target tissue. PCR products are used to construct Illumina sequencing libraries (Supplementary Figure S2A shows more detail).
In summary, the splicing-based RNA tagging (SRT) method provides a convenient and robust tool to profile trans-spliced genes and identify novel transcripts in a tissue-specific manner, with a low false positive rate.