Tag: rna meaning
The function of a large fraction of the human proteome remains poorly characterized. Tagging proteins with a functional sequence is a powerful way to access function, and inserting tags at endogenous genomic loci allows for the preservation of a near-native cellular background. To characterize the cellular role of human proteins systematically and in a native context, we developed a method for tagging endogenous human proteins with GFP that is fast and easily applicable on a genome-wide scale. Our approach allows studying both the location and the interaction partners of the target protein. Our results pave the way for the large-scale generation of endogenously labeled human cell lines for routine functional interrogation of the human proteome.
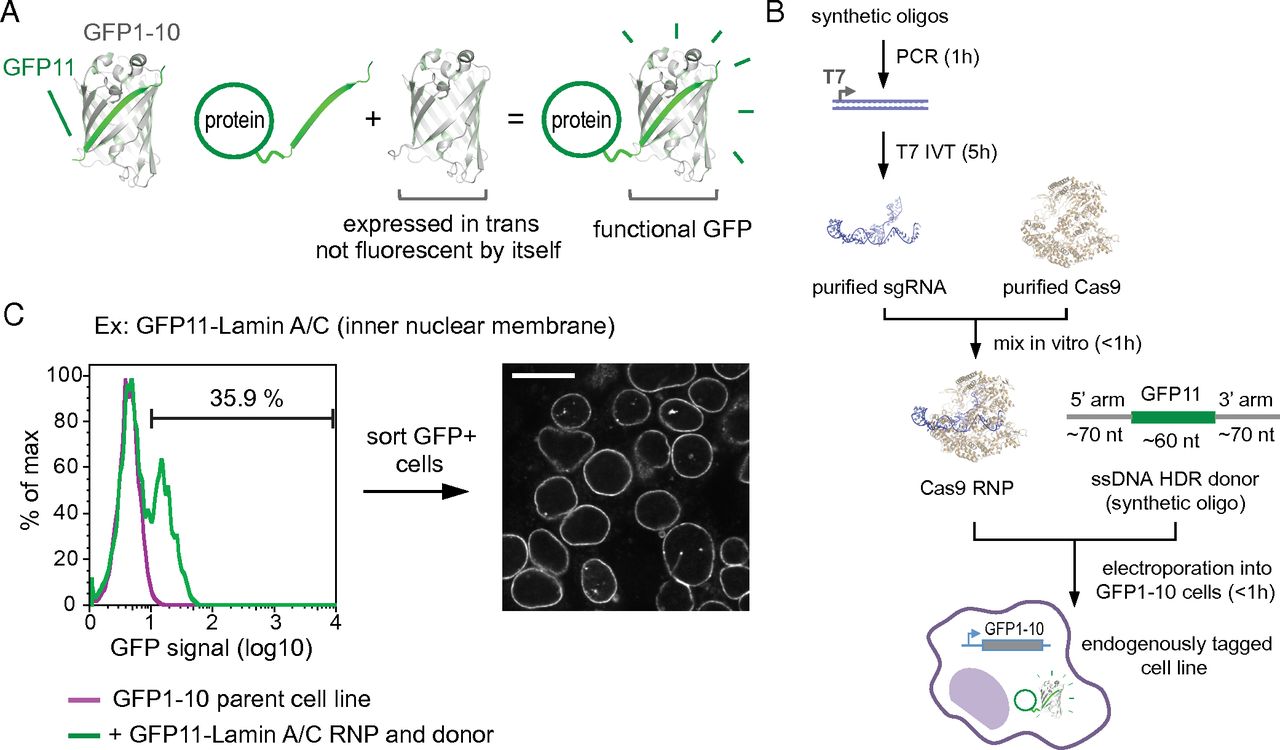
Abstract
A central challenge of the post-genomic era is to comprehensively characterize the cellular role of the approximately 20,000 proteins encoded in the human genome. To systematically study protein function in a native cell background, libraries of human cell lines expressing proteins tagged with a functional sequence at their endogenous loci would be of great value. Here, using single guide RNA ribonucleoprotein / Cas9 nuclease electroporation and leveraging a split GFP system, we describe a scalable method for robust, scarless, and specific labeling of endogenous human genes with GFP. Our approach does not require molecular cloning and allows a large number of cell lines to be processed in parallel. We demonstrate the scalability of our method by targeting 48 human genes and demonstrate that the resulting GFP fluorescence correlates with protein expression levels. Here we present how our protocols can be easily adapted for tagging a given target with GFP repeats, critically enabling the study of low abundance proteins. Finally, we show that our GFP labeling approach allows the biochemical isolation of native protein complexes for proteomic studies. Taken together, our results pave the way for the large-scale generation of endogenously labeled human cell lines for whole-proteome analysis of protein localization and interaction networks in a native cellular context.