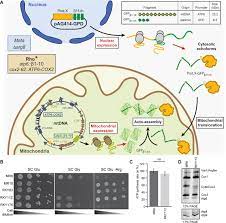
A single nuclear gene can be translated into a dual-localized protein that is distributed between the cytosol and the mitochondria. Accumulating evidence shows that mitoproteomes contain many of these dual localized proteins called ecoforms. Unraveling the existence of mitochondrial ecoforms using current GFP (green fluorescent protein) fusion microscopy approaches is extremely difficult because the GFP signal from the cytosolic ecoform will almost inevitably mask that of the mitochondrial ecoform. Therefore, we designed a yeast strain that expresses a new type of Split-GFP that we call Bi-Genomic Mitochondrial-Split-GFP (BiG Mito-Split-GFP). Because a fraction of the GFP is translated from the mitochondrial machinery while the other is fused with the protein of interest encoded in the nucleus that is translated in the cytosol, the self-assembly of this bi-genomically encoded Split-GFP is limited to mitochondria. We could authenticate the mitochondrial importability of any yeast protein or ecoform, but also of other organisms such as the human Argonaute 2 mitochondrial ecoform.
Introduction
Mitochondria provide aerobic eukaryotes adenosine triphosphate (ATP), which involves the oxidation of carbohydrates and fatty acids (Saraste, 1999), as well as many other vital functions such as the synthesis of lipids and sterols (Horvath and Daum, 2013) and the formation of an iron-sulfur group. (Lill et al., 2012). Mitochondria have their own genome, a remnant of an ancestral prokaryotic genome (Gray, 2017; Margulis, 1975) that has been considerably reduced in size due to massive gene transfer during eukaryotic evolution (Thorsness and Weber, 1996). As a result, most of the proteins necessary for mitochondrial structure and functions are expressed from the nuclear genome (> 99%) and synthesized as mitochondrial-targeted precursors by mitochondrial targeting signals (MTS), which in some cases they split on import (Chacinska et al., 2009). In yeast S. cerevisiae, it has been suggested that approximately one third of the mitochondrial proteins (mitoproteome) are dual-localized (Ben-Menachem et al., 2011; Dinur-Mills et al., 2008; Kisslov et al., 2014 ), and have been called ecoforms (or ecoproteins) to accentuate the fact that two identical or nearly identical forms of a protein can reside in the mitochondria and in another compartment (Ben-Menachem & Pines, 2017). Due to these two coexisting forms and the difficulty in obtaining pure mitochondria, determining a complete mitoproteome remains a challenge and led to conflicting results (Kumar et al., 2002; Morgenstern et al., 2017; Reinders et al., 2006; Sickmann et al., 2003).
Among all the possible methods used to identify the subcellular fate of a protein, engineering of green fluorescent protein (GFP) fusions has the main advantage that these fusions can be visualized in living cells using epifluorescence microscopy. This method is suitable for discriminating the cytosolic and mitochondrial groups of dual-localized proteins when the cytosolic fraction has a lower concentration than the mitochondrial one (Weill et al., 2018). However, when the cytosolic ecoform is more abundant than the mitochondrial, this will inevitably overshadow the mitochondrial fluorescence signal. To avoid this drawback, we designed a yeast strain that contains a new type of Split-GFP system called Bi-Genomic Mitochondrial-Split-GFP (BiG Mito-Split-GFP) because a fraction of GFP is encoded by the mitochondrial genome, while the other is fused with the protein encoded in the nucleus to be tested. In doing so, both Split-GFP fragments are translated into separate compartments and only mitochondrial proteins or dual localized protein ecoforms trigger GFP reconstitution and can be visualized by live cell fluorescence microscopy.
In this paper, we first validate this system with proteins exclusively localized in the mitochondria and with the dual localized glutamyl-tRNA synthetase (cERS) that resides and functions in both the cytosol and the mitochondria as we have previously demonstrated (Frechin et al., 2009 ; Frechin et al., 2014). Next, we apply our Split-GFP strategy to the almost complete set of all known yeast cytosolic aminoacyl-tRNA synthetases. Interestingly, we discovered that two of them, cytosolic phenylalanyl-tRNA synthetase 2 (cFRS2) and cytosolic histidinyl-tRNA synthetase have a dual location. We also confirm the recently reported dual cell location of cytosolic cysteinyl-tRNA synthetase (cCRS) (Nishimura et al., 2019). Furthermore, we demonstrated that our yeast strain BiG Mito-S