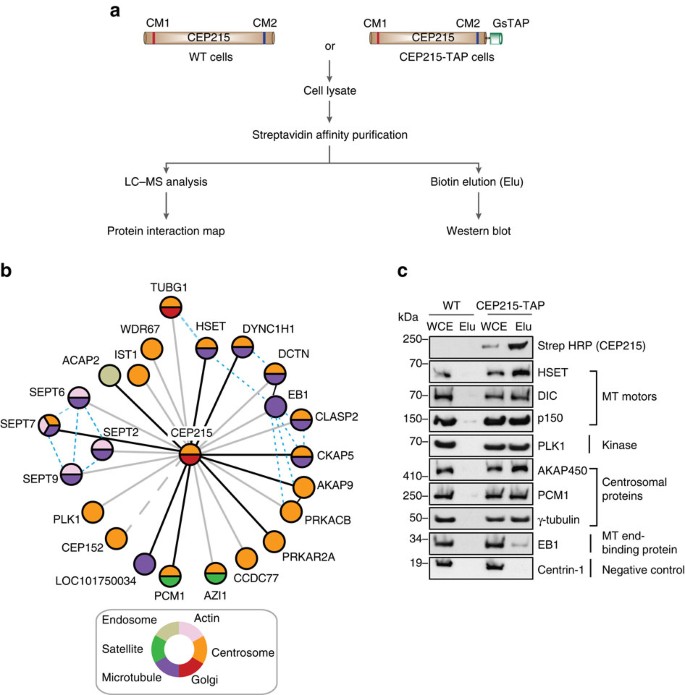
Numerical aberrations of the centrosome underlie certain developmental abnormalities and can promote cancer. A cell maintains a normal centrosome number by coupling the centrosome duplication with segregation, which is achieved by the sustained association of each centrosome with one pole of the mitotic spindle. Although the centrosomal protein linked to microcephaly and primordial dwarfism CEP215 has been implicated in this process, the molecular mechanism responsible remains unclear.
Here, using proteomic profiling, we identify the less end-targeted microtubule motor protein HSET as a direct CEP215 binding partner. The targeted deletion of the HSET-binding domain of CEP215 in vertebrate cells causes detachment of the centrosome and results in the depletion of HSET in the centrosomes, a phenotype also observed in cells derived from CEP215-deficient patients. Furthermore, in cancer cells with centrosome amplification, the CEP215-HSET complex promotes the grouping of additional centrosomes into pseudo-bipolar spindles, ensuring viable cell division. Thus, stabilization of the centrosome-spindle pole interface by the CEP215-HSET complex could promote the survival of cancer cells containing supernumerary centrosomes.
Introduction
Centrosomes act as the dominant microtubule assembly sites in mitosis and, therefore, the number of centrosomes corresponds to the number of spindle poles formed1. Because the faithful transmission of genetic information requires a bipolar mitotic spindle, the number of centrosomes must be strictly controlled in the cells. Consequently, centrosome numbers are regulated by two mechanisms. First, the duplication of the centrosome is limited to once per cell cycle, ensuring that cells enter mitosis with two functional centrosomes2,3. Second, each centrosome associates and co-secretes with its own pole of the mitotic spindle, which causes each daughter cell to inherit precisely one centrosome4. The centrosomes and the mitotic spindle poles are distinct structures, well illustrated by the presence of focused spindle poles in cells that lack centrosomes5,6,7. Spindle pole formation relies on microtubule motors and microtubule-associated proteins that cross over and focus kinetochore-associated microtubule bundles (k-fibers).
In Drosophila S2 cells, the key protein responsible for maintaining the centrosomes at the spindle poles is dynein, a motor directed to the negative end8,9,10. Dynactin increases the processivity of dynein and together they transport the integrity protein from the spindle pole, the nuclear mitotic apparatus (NuMA), to the negative ends of the spindle microtubules11,12. In NuMA-deficient mammalian cells, the k fibers lose focus and the centrosomes detach from the poles13. Similar phenotypes have been documented in Drosophila cells and embryos after disruption of the less end-directed, non-claret-disjunctional (ncd) motor protein kinesin-14 (ncd) 10,14.
In contrast, the mammalian homologous HSET is largely dispensable for the k-fiber approach. Instead, HSET contributes to spindle elongation through crosslinking and sliding microtubules, functions that depend on its C-terminal motor domain and the additional microtubule-binding site on its N-terminal tail. Both ncd and HSET have been implicated in the survival of cells with amplification of the centrosome16,17,18,19. In particular, orthologs mediate the clustering of supernumerary centrosomes into pseudo-bipolar spindles, an essential function for the continued proliferation of cells with centrosome amplification. HSET also promotes acentrosomal spindle pole clustering17.
The centrosome comprises a pair of centrioles embedded in the pericentriolar matrix (PCM), the nucleation site of microtubules. CEP215 is an evolutionarily conserved PCM protein present in microtubule organization centers from yeast to humans; the centrosomin 1 (CM1) motif at its N end binds to the γ-tubulin complex20,21,22,23. CEP215 organizes various PCM components, including pericentrin and AKAP450 (refs 24, 25, 26, 27, 28, 29, 30). The deletion of centrosomin (cnn), its Drosophila ortholog, disruption of the CM1 domain of chicken CEP215, and depletion of CEP215 in HeLa cells cause centrosome shedding from the mitotic spindle poles27,31,32. However, the focus of the spindle pole remains in cells deficient in CM1, consistent with the normal localization of NuMA and dynactin27. Mutations in CEP215 are associated with congenital diseases such as primary microcephaly and primordial dwarfism33,34.
Here we set out to identify the molecular mechanism