RNA scanning enables researchers to visualize RNA molecules in cells and tissues, providing important spatio-temporal information on RNA dynamics and function. Methods such as fluorescent in situ hybridization (FISH) and molecular beacons rely on complementary oligonucleotides to label and view endogenous transcripts. Other methods create artificial chimeric transcripts coupled with bacteriophage-derived coat proteins (eg, MS2, λN) to label molecules in living cells. In other approaches, endogenous RNAs are recognized by complementary RNAs complexed with non-catalytic Cas proteins. Each technique has its own set of strengths and limitations that must be considered when planning an experiment. Here, we discuss the mechanisms, advantages, and weaknesses of in situ hybridization, molecular beacons, MS2 tagging, and Cas-derived systems, as well as how RNA screening can be employed to study various aspects of molecular biology.
1. Introduction
RNA molecules have a wide range of functions in the cell. Coding RNAs (messenger RNA (m)) serve as templates for protein translation, while noncoding RNAs (including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) regulate gene expression programs in many levels [1, 2]. RNAs govern all aspects of cellular metabolism and therefore have been shown to be essential regulators of physiological and pathological processes [3, 4].
Recent advances in RNA biotechnology have allowed researchers to interrogate the transcriptome en masse. Microarrays are rapid and cost-effective methods for quantifying RNA levels in different populations, and RNA sequencing (RNA-seq) can further elucidate the identity of RNAs at the cellular or tissue level [5-7]. Immunoprecipitation (IP) of native or cross-linked ribonucleoprotein (RNP) complexes (RIP and CLIP, respectively) can further identify the targets of a given RNA-binding protein and provide information on RNA-protein interactions [8] . These methods have transformed the field of RNA. However, none of these techniques provide information about the spatial or temporal dynamics of the RNA in the cell, including its location as the RNA is processed, transported, stored, translated, or degraded. To study these parameters, researchers must somehow label an RNA of interest and analyze it using microscopy.
Advances in RNA localization at the subcellular level have improved our understanding of viral and neurodegenerative diseases by characterizing RNA-mediated pathological mechanisms. For example, analysis of hepatitis C virus (HCV) RNA trafficking revealed that host cells increase the production of type I interferons by packaging viral RNAs into exosomes; these are then transported to nearby dendritic cells, activating the antiviral response mediated by TLR-7 [9]. Other studies revealed that RNA export from the nucleus was inhibited by the formation of toxic protein aggregates [10], an observation that may be relevant for Alzheimer's and Huntington's diseases, for example, as they are driven by the accumulation of toxic proteins [11]. .
RNA localization is also important in establishing cell polarity and asymmetry during development. The developmental pattern is achieved by restricting the translation of the mRNA to one side of the cell, effectively localizing the respective protein products in the subcellular compartments. This is evident in Drosophila oocytes, where regulation of bicoid mRNA localization establishes the head and thorax regions in the egg [12]. Similar mechanisms have also been identified in the development of Xenopus and zebrafish [13, 14].
In adult mammals, a notable example of long-range mRNA transport is found in neurons in the form of messenger ribonucleoprotein (mRNP) granules [15]. These mRNP granules are stored in "translation hot spots" within neurons [16]. Neural depolarization triggers mobilization of storage granules to form actively translating polysomes [17]. The complex biological functions of mRNP complexes have been studied for the past 20 years, revealing important links with neuronal function and survival [15, 18, 19].
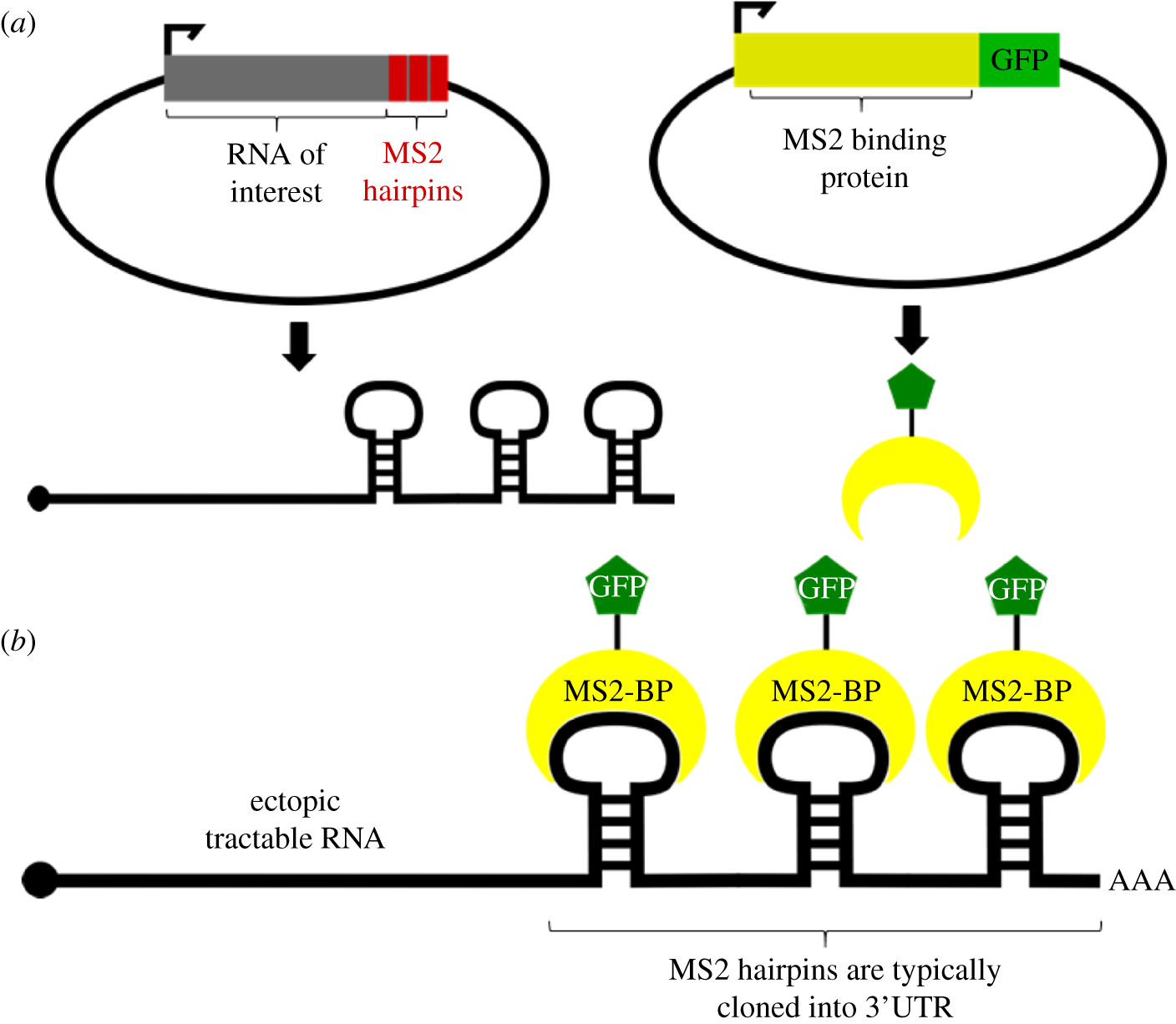
To study the localization and transport of cellular RNA, several RNA screening methods have been developed that have revolutionized the field. This review describes the most popular of these techniques; Specifically, we discuss methodologies for tagging endogenous RNAs using oligomeric tags fluo