Principle of the bead binding assay for soluble proteins. A protein of interest is flanked by a 6His tag at its N-terminus and the GFP S11 tag (b -trand 11, residues 215-230) at its C-terminus through a flexible linker (L). The complementary GFP 1-10 detector fragment (β-strands 1-10, residues 1-214) is co-expressed separately in the same cell (top). The GFP S11 tag associates rapidly with GFP 1-10, committing the GFP to fold and form the fluorophore, whether the protein of interest subsequently remains soluble (left) or aggregates (right). Colonies resting on Durapore membranes w are.
We describe an in vitro colony screening to identify Escherichia coli expressing soluble proteins and assembled and stable multiprotein complexes. Proteins with an N-terminal 6His tag and a C-terminal green fluorescent protein (GFP) S11 tag are fluorescently labeled in cells by complementation with a co-expressed 1-10 GFP fragment. After the partial colony lysi.
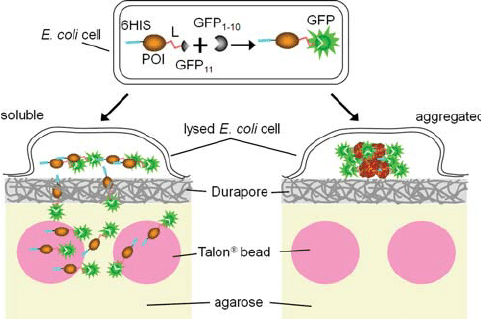
.. chemical lysis of colonies can lead to cross-contamination of clones during subsequent harvesting. Whole cell fluorescence is proportional to the level of expression of GFP S11-labeled proteins when co-expressed with GFP 1-10 (Cabantous et al., 2005). In the immobilized bead assay, we plan to use colony fluorescence to monitor the expression level of the test protein. Since the next step in the assay is to lyse the cells and measure the soluble protein released by fluorescence (Complementary Fig. S3), it is important that the solubility of the labeled protein is as similar as possible to the unlabeled protein. In previous work, we showed that fusion with intact GFP can substantially reduce the solubility of proteins compared to the expression of proteins alone (P ́delacq et al., 2006).
Since the co-expression of GFP 1-10 and S11-tagged proteins leads to a fraction of fused GFP, we wondered if this could strongly disturb the solubility of the protein. In earlier work, we lysed cells expressing GFP S11-tagged proteins and then added GFP 1-10 to clarified lysates to quantify soluble protein (Cabantous et al., 2005). We asked whether GFP S11-tagged proteins behave differently after lysis if they have previously complemented in the cell with GFP 1-10. To study the effect of GFP 1-10 co-expression on the solubility of GFP S11 proteins in greater detail, we performed in vitro split GFP assays on the soluble and sediment fractions of 18 control proteins (Materials and methods) expressed alone or with GFP S11 tags, as described (Cabantous et al., 2005; Cabantous and Waldo, 2006), and the fraction of protein expressed in soluble form tabulated (Table I).
In a separate experiment, we coexpressed the 18 control proteins with GFP S11 tags together with GFP 1-10 in liquid culture and calculated the fraction of fluorescent protein that was soluble (Materials and methods, Table I). We also measured the Talon w-bound fluorescence for further testing (see Assay Bead Assay; comparison with liquid culture below). The fraction of each protein expressed in soluble form was well correlated for the two expression methods (linear correlation coefficient R 2 1⁄4 0.88, Complementary Fig. S4). We concluded that, unlike direct GFP fusions, GFP labeling of proteins using the split GFP co-expression protocol did not strongly disturb protein solubility. This is probably because the S11 tag was designed not to disturb the solubility of the protein, and that during co-expression, the S11 tagged protein has the ability to substantially complete its folding on the ribosome before interacting with the GFP 1 fragment. -10 in trans. . Furthermore, GFP 1-10 was also designed not to aggregate or complete its folding before interacting with the S11 tag, further reducing the likelihood that GFP 1-10 will interfere with the folding of the target protein. In direct GFP fusions, the upstream protein can interfere with the folding of the fused GFP domain (Waldo et al., 1999; P ́delacq et al., 2002). Sonication is not feasible for the colony-based assay. We previously showed that chemical lysis using SoluLyse w gave sonication-like solubility for a library of acyl carrier protein domain fragments (Listwan et al., 2010).
To study the effect of buffer conditions in SoluLyse w on the solubility of a set of proteins, we compared sonication in SoluLyse w with sonication in TNG buffer for a set of 18 control proteins (Materials and methods). The cultures expressing both GFP 1-10 and S11-labeled proteins were lysed by sonication in the optimized SoluLyse w cocktail (Materials and methods, Optimization of the lysis cocktail and conditions for 18 controls) or in TNG, and then the soluble, insoluble and Talon w each of the bound fluorescences were measured (Table I). The solubility (soluble protein divided by total protein) of each co-induced sample sonicated in the presence of SoluLyse w was compared to the corresponding solubility obtained using TNG sonication (Fig.