Tag: isotopes of nitrogen
RNAs are key players in life, as they connect the genetic code (DNA) with all cellular processes dominated by proteins. They contain a variety of chemical modifications, and many RNAs fold into complex structures. Here, we review recent progress in RNA structure and modification analysis based on stable isotope labeling techniques. Mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR) are the key tools and many advances were made possible by the analysis of stable isotope-labeled RNA. Therefore, we discuss current stable isotope labeling techniques such as metabolic labeling, enzyme labeling, and chemical synthesis. Analysis of RNA structure by NMR is challenging due to two major problems that become even more important when RNA size increases, namely, chemical shift overlaps and line broadening leading to complete loss of signal. . Various isotope labeling strategies have been developed to provide solutions to these important problems, such as deuteration, segmental isotope labeling, or site-specific labeling. Quantification of modified nucleosides in RNA by MS is only possible by applying internal standards labeled with stable isotopes. With mass spectrometry coupled to nucleic acid isotope labeling (NAIL-MS), it is now possible to analyze the dynamic modification and demodification processes of post-transcriptional RNA. The trend, both in NMR analysis and MS RNA analysis, is undoubtedly shifting from instantaneous moment analysis to the development and application of tools capable of analyzing the dynamics of RNA structure and modification profiles.
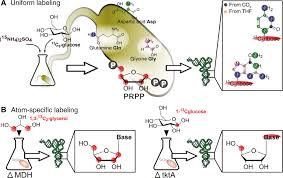
Keywords: epitranscriptomics; mass spectrometry; NAIL-MS; NMR spectroscopy; RNA modifications; RNA structure; segmental labeling; stable isotope labeling
Funding source: DFG
Grant identifier / grant number: KE 1943 / 3-1
Financing statement: S.K. acknowledges funding for the DFG's Emmy-Noether program (Founder's Id: http://dx.doi.org/10.13039/501100001659, KE 1943 / 3-1). P.B. and C.T. We are grateful for the financial support of the CNRS, the ANR TriggeRNA (ANR-08-PCVI-0025), the ANR NMR-VitAmin (ANR-14-CE09-0012-01) and the Labex DYNAMO.
Expressions of gratitude
We thank Matthias Heiß for providing graphic support in the generation of figures.
References
Alvarado, L.J., LeBlanc, R.M., Longhini, A.P., Keane, S.C., Jain, N., Yildiz, Z.F., Tolbert, B.S., D’Souza, V.M., Summers, M.F., Kreutz, C., et al. (2014a). Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates studies of the structure and dynamics of RNA. ChemBioChem 15, 1573-1577. Search in Google Scholar
Alvarado, L.J., Longhini, A.P., LeBlanc, R.M., Chen, B., Kreutz, C. and Dayie, T.K. (2014b). Chemoenzymatic synthesis of RNA selectively labeled with 13C / 15N for NMR structure and dynamics studies. Enzymol methods. 549, 133-162. Search in Google Scholar
Aoyagi, T., Hanada, S., Itoh, H., Sato, Y., Ogata, A., Friedrich, M.W., Kikuchi, Y. and Hori, T. (2015). Ultra-high sensitivity stable isotope probing of rRNA by high-throughput sequencing of isopycnic centrifugal gradients. Reign. Microbiol. Rep. 7, 282-287. Search in Google Scholar
Barnwal, R.P., Loh, E., Godin, K.S., Yip, J., Lavender, H., Tang, C.M. and Varani, G. (2016). Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic Acids Res. 44, 9426-9437. Search in Google Scholar
Barnwal, R.P., Yang, F. and Varani, G. (2017). Applications of NMR for the determination of large and small RNA structures. Bow. Biochem. Biophys. 628, 42–56. Search in Google Scholar
Baronti, L., Karlsson, H., Marusic, M. and Petzold, K. (2018). A guide to large-scale RNA sample preparation. Anal. Bioanal. Chem. 410, 3239-3252. Search in Google Scholar
Barraud, P., Gaudin, C., Dardel, F. and Tisne, C. (2007). New insights into the formation of the HIV-1 reverse transcription initiation complex. Biochimie 89, 1204-1210. Search in Google Scholar
Batey, R.T., Inada, M., Kujawinski, E., Puglisi, J.D. and Williamson, J.R. (1992). Preparation of Isotopically Labeled Ribonucleotides for RNA Multidimensional NMR Spectroscopy. Nucleic Acids Res. 20, 4515–4523. Search in Google Scholar
Batey, R.T., Battiste, J.L. and Williamson, J.R. (nineteen ninety five). Preparation of isotopically enriched RNA for heteronuclear NMR. Enzymol methods. 261, 300–322. Search in Google Scholar
Beusch, I., Barraud, P., Moursy, A., Clery, A. and Allain, F.H. (2017). The hnRNP A tandem RNA recognition motifs