rna sequencing
Bioengineers at EPFL have developed a new method for bulk RNA sequencing that combines the multiplexing-based cost effectiveness of a single cell RNA sequencing workflow with the throughput of a bulk RNA sequencing procedure.
RNA sequencing is a technique used to analyze entire genomes by looking at the expression of their genes. Today, these genome-wide expression analyzes are a standard tool for genomic studies because they are based on high-throughput technologies, which in turn have become widely available.
However, RNA sequencing is still expensive and time consuming, because it first requires the expensive preparation of a complete genomic library, the pool of DNA generated from the RNA of cells, while the data itself is also difficult to determine. analyze. All of this makes it difficult to execute RNA sequencing, making its adoption not as widespread as it could be.
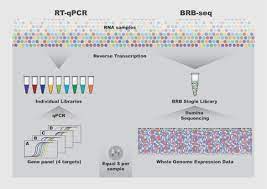
Some new approaches have been introduced to help, driven by the single cell transcriptomics revolution, which uses what is known as "sample barcode" or "multiplexing." Here, individual 'barcode' sequences are added to each DNA fragment during library preparation so that each can be identified and classified prior to final data analysis, meaning this approach only requires a single library containing multiple samples or distinct cells.
Bar codes reduce both cost and time, and this could extend to bulk RNA sequencing of large sets of samples. But there are still problems in adapting and validating protocols for reliable and inexpensive profiling of bulk RNA samples, which is what we are faced with when trying to analyze the cell or tissue transcriptome.
Now scientists in Bart Deplancke's lab at EPFL's Institute for Bioengineering have developed a novel approach called Bulk RNA Bar Sequencing and Coding (BRB-seq) that is 25 times less expensive than a commercial RNA sequencing technology. conventional (Illumina TruSeq).
Among its many advantages, BRB-seq is fast and preserves strand specificity, a challenge in the field, which has to do with transcribing DNA in the right direction. As such, BRB-seq offers a low-cost approach to performing transcriptomics on hundreds of RNA samples, which can increase the number of biological replicas (and therefore experimental precision) in a single run.
In terms of performance, the scientists found that BRB-seq can detect the same number of genes as the "gold standard" in the field, that is, TruSeq stranded mRNA, at the same sequencing depth and that the technique produces data. reliable even with low levels of quality RNA samples. In addition, it generates genome-wide transcriptomic data at a cost comparable to that of four-gene profiling using RT-qPCR, which is currently a standard, but low-throughput, method for measuring gene expression.